
Estefanos Kebebew
Interested at the Intersection of Biology and Code
My Focus
I am a software developer and researcher driven by the challenge of solving complex problems with robust, and scalable code.
I am open for opportunities in both biotech research and development and core software/ML engineering roles, where I can provide unique hybrid skill set to build data-driven solutions.
Research & Projects
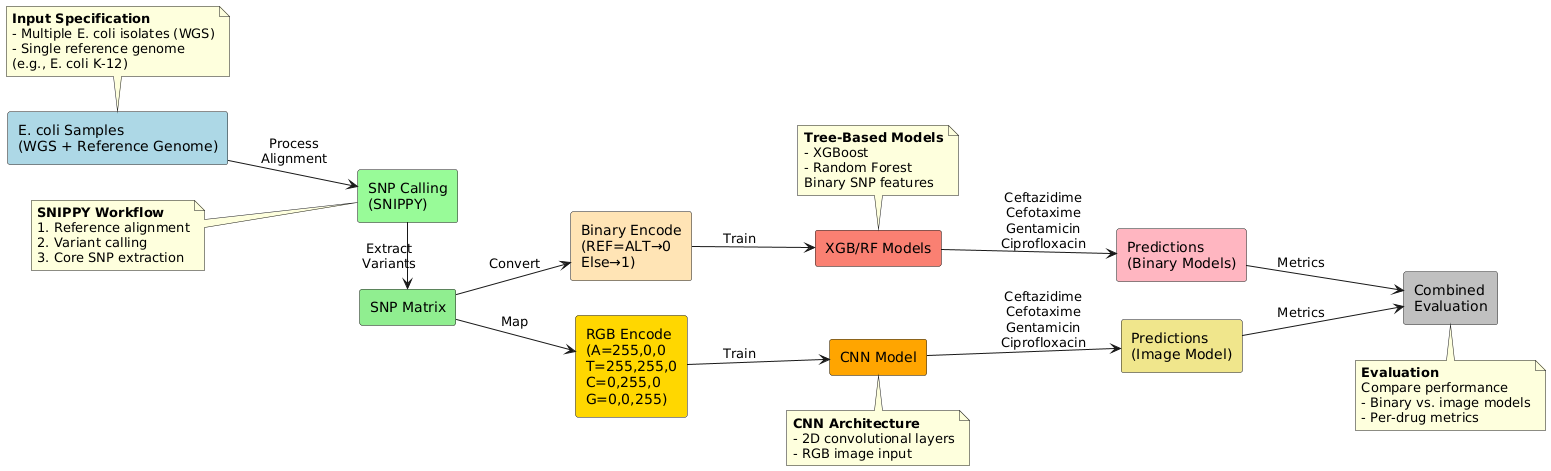
📑 Master's Thesis: Predicting AMR with Deep Learning
- Problem: Traditional lab methods for testing Antimicrobial Resistance (AMR) are slow, delaying crucial treatments.
- Solution: Developed and compared machine learning models (CNNs, Random Forest, XGBoost) to predict AMR in E. coli directly from genomic SNP data.
- Impact: This computational approach aims to reduce prediction time providing rapid, actionable insights for clinical and research settings.
⚙️ Thesis Pipeline
Posters
Technical Skills
🧬 Bioinformatics & Data Science
🤖 ML / Deep Learning
🔥 TensorFlow / PyTorch
📊 Pandas / NumPy
📈 Matplotlib / Seaborn
🧬 Roary / Snippy
🧬 SRA Toolkit
🧬 LDpred / PRSice
🗃️ UK Biobank Data
💻 Software Engineering & DevOps
🐍 Python
☕ Java / Spring
⚛️ React
🗄️ SQL / PostgreSQL
☁️ AWS
🐋 Docker
🐙 Git / GitHub
🐧 Linux / Shell Scripting